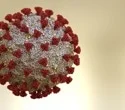
The SARS-CoV-2 XBB.1.16 variant has an effective reproductive number (Re) that is 1.27 and 1.17 times higher than that of the XBB.1 and XBB.1.5 subvariants, respectively, demonstrating the ability of this unique Omicron variant to spread swiftly.
The World Health Organization (WHO) started keeping an eye on XBB.1.16 on March 30, 2023, after it was discovered in various nations, including India, as a result of its heightened transmissibility. Earlier, the F486P alteration in the spike (S) protein of the SARS-CoV-2 Omicron XBB subvariants, including XBB.1.5 and XBB.1.9, were widely disseminated around the world.
The recent investigations revealed that XBB.1.16 had two S substitutions, E180V and T478R in the N-terminal domain (NTD) and receptor-binding domain (RBD), respectively, as compared to its forerunner mutant strains. The dissociation constant (KD) of although XBB.1.16 had a 2.4-fold greater RBD than XBB.1.5 for the host receptor angiotensin-converting enzyme 2 (ACE2), its KD values were noticeably lower than those of XBB.1, indicating that this unique subvariant had lower binding affinities. Pseudovirus experiments revealed that XBB.1.16’s infectivity was similar to that of XBB.1, but different from XBB.1.5, which had a higher infectivity than the parental XBB.1 mutant.
It should be noted that this viral variant’s ability to infect hosts is significantly impacted by the substitution mutations S: T478R and S: E180V. The S: E180V alteration greatly decreased the viral infectivity of XBB.1.16, but the S: T478R mutation increased it
These two S protein mutations were probably acquired simultaneously by XBB.1.16 as part of an evolutionary strategy. Other Omicron subvariants, such as BA.5 and XBB.1, have shown this behavior in the past. In fact, XBB.1.16 has evolved in a manner similar to preceding Omicron subvariants.
Neutralization assays revealed that XBB.1.16 was much more resistant to sera from people who had been reinfected with Omicron BA.2/BA.5, 18 and 37 times more resistant than Omicron B.1.1/B.1.1, in terms of sensitivity to sera. The sensitivity of this variety to convalescent sera of XBB.1-infected hamsters, however, was comparable to that of the XBB.1/XBB.1.5 mutants.
All six clinically available monoclonal antibodies for SARS-CoV-2 were largely ineffective against XBB.1.16. Only sotrovimab, but this impact was minimal, displayed antiviral activity against XBB.1.16. Finally, antigenic mapping revealed that XBB.1.16 had antigenicity similar to XBB.1, but this